1. Introduction
This document describes the openModeller web service interface (OMWS) version 1.0. For more information about the new API version, please check the corresponding OMWS 2.0 documentation. OMWS was created so that the main ecological niche modelling functionalities provided by openModeller (Muñoz et al. 2011) could be available in a standardized way over the web to other programs. Therefore the target audience for this document are developers interested in writing programs to interact with an OMWS instance or perhaps in implementing a new OMWS server. It is assumed that readers have basic understanding about ecological niche modelling, basic understanding of openModeller and knowledge about web services.
This document was originally prepared for the BioVeL project with funds from the European Commission.
2. Technical details
OMWS is based on SOAP following the document/literal style and the interface is programatically defined in WSDL:
http://openmodeller.cria.org.br/ws/1.0/openModeller.wsdl
The main XML elements, attributes, structure and data types used inside the SOAP Body are defined in a separate XML Schema compatible with all openModeller serializable objects:
http://openmodeller.cria.org.br/xml/1.0/openModeller.xsd
3. Software
So far there is only one implementation of the server developed in C++ using gSOAP. This implementation is available as part of the openModeller code under the src/soap/1.0 directory. The server can run as a CGI application, a stand alone server (daemon) or a multi-threaded server. By using gSOAP, stubs are also available for developers willing to write clients in C++ (openModeller Desktop, for example, uses them). There is also a simple command-line client written in Perl that depends on the SOAPLite Perl module. This client is available under the same directory in the openModeller source code (src/soap/1.0/sampleClient.pl). GBIF has some Java code to generate niche models through its portal by interacting with the service: http://code.google.com/p/gbif-dataportal/source/browse/trunk/portal-web/src/main/java/org/gbif/portal/web/controller/openmodeller/
4. Service instances
Currently the following instances are publicly available:
http://modeller.cria.org.br/cgi-bin/om_soap_server.cgi
http://modeller.cria.org.br/ws1/om
Note: you need a SOAP client to interact with these URLs! If you click on the links using your browser you may get an error.
5. Interaction with the service
Although the number of operations available on the service is relatively small, there are many different ways to use them. Figure 1 shows a typical workflow as an example. Some people may be interested only in the models - without even testing of projecting them. Others may want to project a model only if a previous test indicates that the results are good enough. This can actually be done in different ways, since the model result also includes statistics for the training dataset, while "testModel" requests are usually performed with an external dataset. Even model creation may be conditioned by previous tests involving preliminary models created with different subsets of input data (through n-fold cross validation, for instance). Multiple projections can also be requested when there is more than one environmental scenario available.
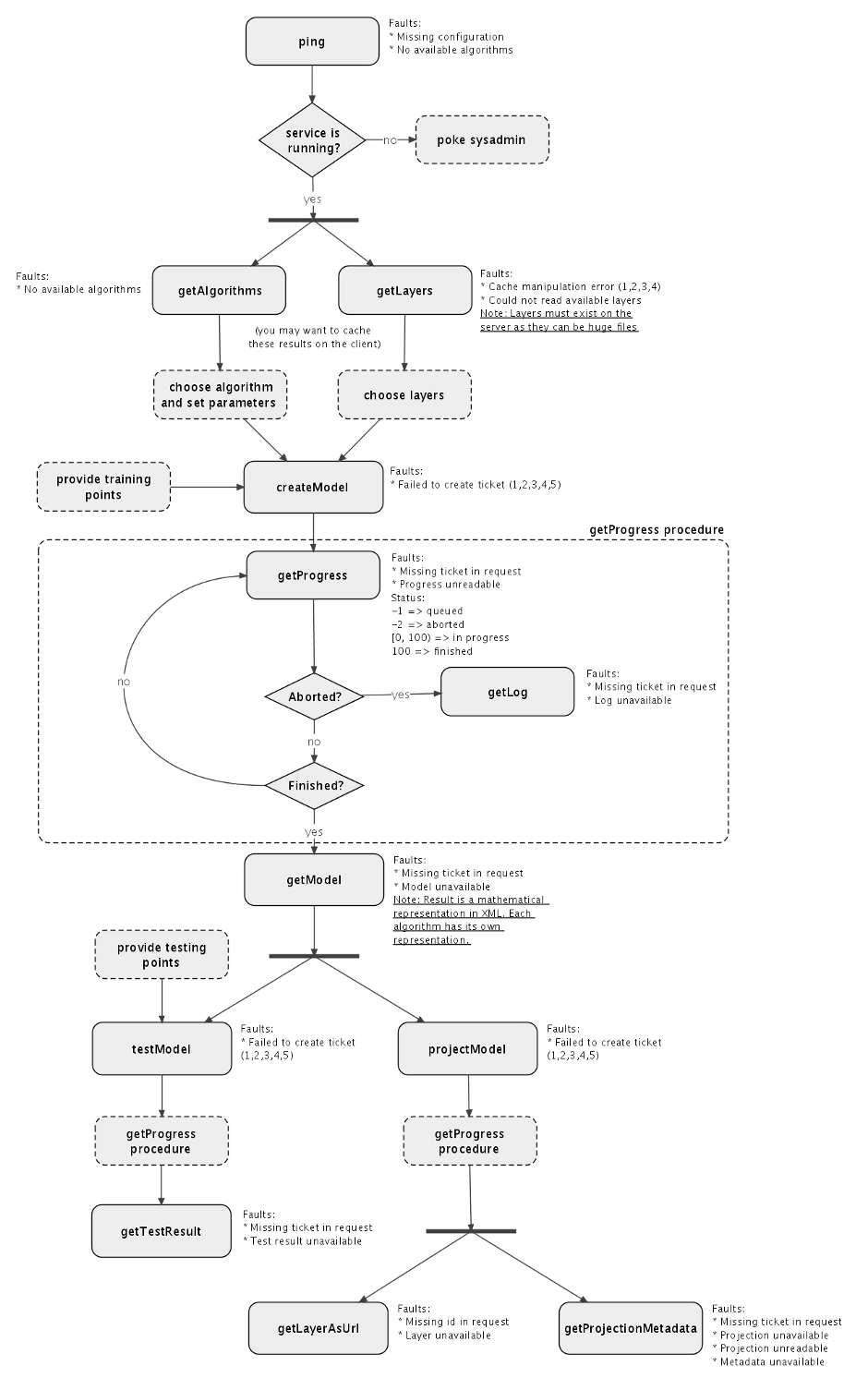
Figure 1: Typical workflow to interact with the service. Boxes with solid lines indicate calls to service operations. Boxes with dashed lines indicate external tasks, except those indicating the whole "getProgress" procedure.
To better understand the functionality, it is important to note that a "model" for openModeller is the mathemathical representation of the ecological niche. If we take a simple algorithm like Bioclim for example, the model will be the bioclimatic envelopes and the logic that says that inside the inner envelope the environmental conditions are suitable (corresponding to the numerical value 1), inside the marginal envelope the environmental conditions can still be considered acceptable (corresponding to 0.5) and outiside the marginal envelope the environmental conditions are unsuitable (corresponding to 0). Therefore, the createModel operation will request the creation of that kind of model - not a map! The result of "getModel" will be the serialized model, which in the case of Bioclim will be just the bioclimatic envelopes (minimum, maximum, average and standard deviation cutoff for each variable). Two things can be done with a model in OMWS: it can be tested as many times as necessary (getting back statistical results as a confusion matrix and/or ROC curve) or it can be projected into a geographic region given an environmental scenario. This can also be done as many times as necessary. The result of a model projection is finally the potential distribution map in the form of a georeferenced raster.
As can be seen, OMWS is centered around a few core operations for ecological niche modelling, not covering other related things such as data cleaning, splitting points into test/training, pre-analysis on input data, post processing distribution maps, etc. Nevertheless, these core operations can still be used and combined in many different ways, keeping the interface simple and effective.
6. Additional notes
All XML examples in this document were extracted from message dumps using the Perl client (through the -debug option) interacting with the http://modeller.cria.org.br/ws1/om service instance running a version of openModeller currently under development on top of the previous 1.2.0 stable version. All XMLs were formatted for better visualization. In some cases they were deliberately shortened to avoid displaying dozens of algorithms and hundreds of layers in the examples. Despite these changes, all XML messages can be seen as real and functional examples of how to interact with the service.
SOAP faults included here are all based on the only existing server implementation and should not be regarded as mandatory functionality for additional server software that may be implemented in the future. Some comments in this document may also be specifically related to the existing server implementation and not just the protocol itself.
7. Operations
7.1. ping
7.1.1. Description
Indicates service availability. In a broader sense, service availability is not just related with connectivity. It also involves correct configuration on the server side. Server implementations can therefore use this method to check as much as possible that the server is prepared to properly handle all requests. Clients must get a <status>1</status> if the service is available - otherwise a SOAP fault is raised. Additionally, server implementations may use the <version> element in the header to indicate their name and version.
This is the right operation for service monitoring.
7.1.2. SOAP faults
- Missing configuration
- No available algorithms
7.1.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:ping xsi:nil="true" xmlns:omws="http://openmodeller.cria.org.br/ws/1.0" />
</soap:Body>
</soap:Envelope>
Example 1: ping request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Header>
<omws:version>oM Server 1.0 (openModeller 1.2.0)</omws:version>
</SOAP-ENV:Header>
<SOAP-ENV:Body>
<omws:pingResponse>
<status>1</status>
</omws:pingResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 2: ping response.
7.2. getAlgorithms
7.2.1. Description
Returns metadata about all modelling algorithms available on the server. Metadata includes id, version, name, overview, description, designers, bibliography, developers, an indicator if the algorithm can handle categorical data, an indicator if it requires absence data, and parameters. Parameter metadata includes id, name, data type (Integer, Real or String), overview, description, accepted range and default value. Algorithm and parameter ids are used to reference them in the other operations.
In the current openModeller version all textual content is in English. When displaying algorithm metadata on user interfaces, descriptions of both algorithms and parameters are particularly important. For an algorithm, this may contain crucial information about how to run it, as in the case of AquaMaps, which requires a particular set of environmental layers to work. In other cases, descriptions may help to understand the differences between two implementations of the same algorithm, such as Desktop GARP and OM GARP, or they may contain additional information about the status of the algorithm and the conditions in which it can be used. For parameters, in many cases descriptions contain the possible values with additional explanations.
An algorithm may have zero, one or more parameters. EnvelopeScore is an example of an algorithm without any parameters. Parameter metadata can optionally indicate an accepted range, with minimum and maximum values (both can be present, or at least one of them).
Different service instances are not guaranteed to provide the same algorithms or the same versions of each algorithm, unless there is a specific policy in this respect controlling the different instances.
7.2.2. SOAP faults
- No available algorithms
7.2.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:getAlgorithms xsi:nil="true" xmlns:omws="http://openmodeller.cria.org.br/ws/1.0" />
</soap:Body>
</soap:Envelope>
Example 3: getAlgorithms request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getAlgorithmsResponse>
<AvailableAlgorithms>
<Algorithms>
<Algorithm Id="SVM" Version="0.4">
<Name>SVM (Support Vector Machines)</Name>
<Overview>Support vector machines (SVMs) are a set of related supervised learning methods that belong to
a family of generalized linear classifiers. They can also be considered a special case of Tikhonov regularization.
A special property of SVMs is that they simultaneously minimize the empirical classification error and maximize the
geometric margin; hence they are also known as maximum margin classifiers. Content retrieved from Wikipedia on the
13th of June, 2007: http://en.wikipedia.org/w/index.php?title=Support_vector_machine&oldid=136646498.
</Overview>
<Description>Support vector machines map input vectors to a higher dimensional space where a maximal
separating hyperplane is constructed. Two parallel hyperplanes are constructed on each side of the hyperplane that
separates the data. The separating hyperplane is the hyperplane that maximises the distance between the two parallel
hyperplanes. An assumption is made that the larger the margin or distance between these parallel hyperplanes the
better the generalisation error of the classifier will be. The model produced by support vector classification only
depends on a subset of the training data, because the cost function for building the model does not care about
training points that lie beyond the margin. Content retrieved from Wikipedia on the 13th of June, 2007:
http://en.wikipedia.org/w/index.php?title=Support_vector_machine&oldid=136646498. The openModeller implementation
of SVMs makes use of the libsvm library version 2.85: Chih-Chung Chang and Chih-Jen Lin, LIBSVM: a library for support
vector machines, 2001. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm.
Release history:
version 0.1: initial release
version 0.2: New parameter to specify the number of pseudo-absences to be generated;
Upgraded to libsvm 2.85;
Fixed memory leaks
</Description>
<Designers>
<Designer Name="Vladimir N. Vapnik"/>
</Designers>
<Bibliography>
1) Vapnik, V. (1995) The Nature of Statistical Learning Theory. SpringerVerlag.
2) Schölkopf, B., Smola, A., Williamson, R. and Bartlett, P.L.(2000). New support vector algorithms. Neural Computation,
12, 1207-1245.
3) Schölkopf, B., Platt, J.C., Shawe-Taylor, J., Smola A.J. and Williamson, R.C. (2001). Estimating the support of a
high-dimensional distribution. Neural Computation, 13, 1443-1471.
4) Cristianini, N. & Shawe-Taylor, J. (2000). An Introduction to Support Vector Machines and other kernel-based learning
methods. Cambridge University Press.
</Bibliography>
<Developers>
<Developer Name="Renato De Giovanni in collaboration with Ana Carolina Lorena"
Contact="renato [at] cria . org . br"/>
</Developers>
<AcceptsCategoricalMaps>0</AcceptsCategoricalMaps>
<RequiresAbsencePoints>0</RequiresAbsencePoints>
<Parameters>
<Parameter Id="SvmType">
<Name>SVM type</Name>
<Type>Integer</Type>
<Overview>Type of SVM: 0 = C-SVC, 1 = Nu-SVC, 2 = one-class SVM</Overview>
<Description>Type of SVM: 0 = C-SVC, 1 = Nu-SVC, 2 = one-class SVM</Description>
<AcceptedRange Min="0" Max="2"/>
<Default>0</Default>
</Parameter>
<Parameter Id="KernelType">
<Name>Kernel type</Name>
<Type>Integer</Type>
<Overview>Type of kernel function: 0 = linear: u'*v , 1 = polynomial: (gamma*u'*v + coef0)^degree ,
2 = radial basis function: exp(-gamma*|u-v|^2)
</Overview>
<Description>Type of kernel function: 0 = linear: u'*v , 1 = polynomial: (gamma*u'*v + coef0)^degree ,
2 = radial basis function: exp(-gamma*|u-v|^2)</Description>
<AcceptedRange Min="0" Max="4"/>
<Default>2</Default>
</Parameter>
<Parameter Id="Degree">
<Name>Degree</Name>
<Type>Integer</Type>
<Overview>Degree in kernel function (only for polynomial kernels).</Overview>
<Description>Degree in kernel function (only for polynomial kernels).</Description>
<AcceptedRange Min="0"/>
<Default>3</Default>
</Parameter>
<Parameter Id="Gamma">
<Name>Gamma</Name>
<Type>Real</Type>
<Overview>Gamma in kernel function (only for polynomial and radial basis kernels). When set to zero, the
default value will actually be 1/k, where k is the number of layers.</Overview>
<Description>Gamma in kernel function (only for polynomial and radial basis kernels). When set to zero, the
default value will actually be 1/k, where k is the number of layers.</Description>
<Default>0</Default>
</Parameter>
<Parameter Id="Coef0">
<Name>Coef0</Name>
<Type>Real</Type>
<Overview>Coef0 in kernel function (only for polynomial kernels).</Overview>
<Description>Coef0 in kernel function (only for polynomial kernels).</Description>
<Default>0</Default>
</Parameter>
<Parameter Id="C">
<Name>Cost</Name>
<Type>Real</Type>
<Overview>Cost (only for C-SVC types).</Overview>
<Description>Cost (only for C-SVC types).</Description>
<AcceptedRange Min="0.001"/>
<Default>1</Default>
</Parameter>
<Parameter Id="Nu">
<Name>Nu</Name>
<Type>Real</Type>
<Overview>Nu (only for Nu-SVC and one-class SVM).</Overview>
<Description>Nu (only for Nu-SVC and one-class SVM).</Description>
<AcceptedRange Min="0.001" Max="1"/>
<Default>0.5</Default>
</Parameter>
<Parameter Id="ProbabilisticOutput">
<Name>Probabilistic output</Name>
<Type>Integer</Type>
<Overview>Indicates if the output should be a probability instead of a binary response (only available for
C-SVC and Nu-SVC).</Overview>
<Description>Indicates if the output should be a probability instead of a binary response (only available for
C-SVC and Nu-SVC).</Description>
<AcceptedRange Min="0" Max="1"/>
<Default>1</Default>
</Parameter>
<Parameter Id="NumberOfPseudoAbsences">
<Name>Number of pseudo-absences</Name>
<Type>Integer</Type>
<Overview>Number of pseudo-absences to be generated (only for C-SVC and Nu-SVC when no absences have
been provided). When absences are needed, a zero parameter will default to the same number of presences.</Overview>
<Description>Number of pseudo-absences to be generated (only for C-SVC and Nu-SVC when no absences have
been provided). When absences are needed, a zero parameter will default to the same number of presences.</Description>
<AcceptedRange Min="0"/>
<Default>0</Default>
</Parameter>
</Parameters>
</Algorithm>
<Algorithm Id="ENVSCORE" Version="0.1">
<Name>Envelope Score</Name>
<Overview>Uses the minimum and maximum observed value for each environmental variable to define bioclimatic
envelopes. Probability of occurrence for a point is determined by the number of environmental variables within which the
environmental values at the point fall into the min/max criteria.</Overview>
<Description>Implements a lax Bioclimatic Envelope Algorithm. For each given environmental variable the
algorithm finds the minimum and maximum at all occurrence sites. During model projection, the probability of occurrences
is determined as:
p = layers within min-max threshold / number of layers
Thus choosing a threshold of p=1 gives you the same model output as the original Bioclim model taking both the Suitable
& Marginal classes as predicted presences. The Envelope Score algorithm is equivalent to the inclusive 'OR' implementation
of Bioclim described in Pineiro et al (2007).
</Description>
<Designers>
<Designer Name="Nix, H. A."/>
</Designers>
<Bibliography>Nix, H.A. (1986) A biogeographic analysis of Australian elapid snakes. In: Atlas of Elapid Snakes of
Australia. (Ed.) R. Longmore, pp. 4-15. Australian Flora and Fauna Series Number 7. Australian Government Publishing Service:
Canberra.
Piñeiro, R., Aguilar, J. F., Munt, D. D. & Feliner, G. N. (2007) Ecology matters: Atlantic-Mediterranean disjunction
in the sand-dune shrub Armeria pungens (Plumbaginaceae). Molecular Ecology. 16, 2155-2171.
</Bibliography>
<Developers>
<Developer Name="Tim Sutton and Chris Yesson" Contact="tim [at] linfiniti.com"/>
</Developers>
<AcceptsCategoricalMaps>0</AcceptsCategoricalMaps>
<RequiresAbsencePoints>0</RequiresAbsencePoints>
<Parameters/>
</Algorithm>
</Algorithms>
</AvailableAlgorithms>
</omws:getAlgorithmsResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 4: getAlgorithms response.
7.3. getLayers
7.3.1. Description
Returns all environmental layers available on the server side. Layers can be grouped and organized in a hierarchical structure by means of <LayersGroup> elements. Each layer group and each individual layer has an id and a label. Ids are used to reference layers in the other operations. An optional layer attribute (HasProjection) may indicate that there is no geographical projection information available for the layer. In such cases, openModeller will assume a latlong projection with the WGS84 datum when reading data from the corresponding layer.
Historically, the openModeller server implementation only worked with layers that were locally accessible (i.e., stored on the server or at least accessible from the server through the file system). However, recent versions do allow remote layers that are accessible through the WCS protocol to be used. In this case, the layer id should follow the pattern:
wcs>wcs_service_url>coverage_id
The protocol does not include operations to manipulate layers (add or remove files, or edit metadata). These tasks need to be performed on the server side by other means.
Different service instances are not guaranteed to provide the same layers, unless there is a specific policy in this respect controlling the different instances. Additionally, a service instance may not keep the same layer structure or even the identifiers for all layers over the time - again, unless there is a specific policy with this purpose.
7.3.2. SOAP faults
- Cache manipulation error
- Could not read available layers
7.3.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:getLayers xmlns:omws="http://openmodeller.cria.org.br/ws/1.0" />
</soap:Body>
</soap:Envelope>
Example 5: getLayers request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getLayersResponse>
<AvailableLayers xmlns="http://openmodeller.cria.org.br/xml/1.0">
<LayersGroup Id="1"><Label>layers</Label>
<LayersGroup Id="2"><Label>climate</Label>
<LayersGroup Id="3"><Label>terrestrial</Label>
<LayersGroup Id="4"><Label>global</Label>
<LayersGroup Id="5"><Label>worldclim</Label>
<LayersGroup Id="6"><Label>present</Label>
<LayersGroup Id="7"><Label>bioclim</Label>
<LayersGroup Id="8"><Label>10arc-minutes</Label>
<Layer Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66" HasProjection="0">
<Label>bio 01 - Annual Mean Temperature</Label></Layer>
<Layer Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/bf441530-04d9-11e1-be50-0800200c9a66" HasProjection="0">
<Label>bio 02 - Mean Diurnal Range (Mean of monthly (max temp - min temp))</Label></Layer>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
<LayersGroup Id="9"><Label>marine</Label>
<LayersGroup Id="10"><Label>global</Label>
<LayersGroup Id="11"><Label>incofish</Label>
<LayersGroup Id="12"><Label>HCAFv4</Label>
<LayersGroup Id="13"><Label>present</Label>
<LayersGroup Id="14"><Label>30arc-minutes</Label>
<Layer Id="/layers/en/marine/global/incofish/HCAFv4/present/30arc-minutes/61e7a060-04bf-11e1-be50-0800200c9a66" HasProjection="0">
<Label>Mean annual sea ice concentration</Label></Layer>
<Layer Id="/layers/en/marine/global/incofish/HCAFv4/present/30arc-minutes/9602dc20-04bf-11e1-be50-0800200c9a66" HasProjection="0">
<Label>Mean annual primary production (chlorophyll A) in mgC per square meter per day</Label></Layer>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</LayersGroup>
</AvailableLayers>
</omws:getLayersResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 6: getLayers response.
7.4. createModel
7.4.1. Description
Starts a new model creation job. This requires a set of layers referenced by their ids, a mask (which is also a layer!) referenced by its id, a set of presence points (samples where the environment is suitable for the species), an optional set of absence points (samples where the environment is unsuitable for the species), a modelling algorithm referenced by its id and, if the algorithm requires parameters, a set of parameters referenced by ids and followed by a value.
Each set of points needs a spatial reference system specified in WKT. Example 7 shows the WKT for latlong in WGS84. Longitude and latitude are represented by X and Y, respectively, and decimals must always be separated by a dot. An identification for each point is highly recommended, as points are referenced by it in the logs. The label for each set of points can be used to store the species name. This information is not used by the server, but it is returned later as part of the model created and can be a way to keep record of which species was modelled.
The mask is used to discard points outside its extent or inside cells with nodata.
An optional section <Options> can be used to instruct the server to apply filters in the points before they are used. <SpatiallyUnique/> is used to discard duplicate points (same coordinates) and <EnvironmentallyUnique/> is used to discard points with the same environmental conditions.
An optional <Statistics> section can be used to specify statistics parameters for an internal test (a test with the training data). This section is described in the testModel operation. By default and for backwards compatibility reasons, if this section is not present, the service will calculate statistics with default values. Statistics results are also explained in the testModel operation.
This operation is asynchronous. It immediately returns a ticket that must be used to track job progress later.
7.4.2. SOAP faults
- Failed to create ticket
7.4.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<SOAP-ENV:Envelope xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
SOAP-ENV:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/">
<SOAP-ENV:Body>
<omws:createModel xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
SOAP-ENV:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ModelParameters xmlns="http://openmodeller.cria.org.br/xml/1.0">
<Sampler>
<Environment>
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66" IsCategorical="0"/>
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/bf441530-04d9-11e1-be50-0800200c9a66" IsCategorical="0"/>
<Mask Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66"/>
</Environment>
<Presence Label="Test species">
<CoordinateSystem>GEOGCS['1924 ellipsoid',DATUM['Not_specified',SPHEROID['International 1924',6378388,297,
AUTHORITY['EPSG','7022']],AUTHORITY['EPSG','6022']],PRIMEM['Greenwich',0,AUTHORITY['EPSG','8901']],UNIT['degree',
0.0174532925199433,AUTHORITY['EPSG','9108']],AUTHORITY['EPSG','4022']]</CoordinateSystem>
<Point Id="1" X="-68.85" Y="-11.15"/>
<Point Id="2" X="-67.38" Y="-14.32"/>
<Point Id="3" X="-67.15" Y="-15.52"/>
<Point Id="4" X="-65.12" Y="-16.73"/>
<Point Id="5" X="-63.17" Y="-17.8"/>
</Presence>
</Sampler>
<Algorithm Version="0.2" Id="BIOCLIM">
<Parameters>
<Parameter Value="0.8" Id="StandardDeviationCutoff"/>
</Parameters>
</Algorithm>
<Options>
<OccurrencesFilter>
<SpatiallyUnique/>
<EnvironmentallyUnique/>
</OccurrencesFilter>
</Options>
</ModelParameters>
</omws:createModel>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 7: createModel request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:createModelResponse>
<ticket>FXPAHp</ticket>
</omws:createModelResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 8: createModel response.
7.5. getProgress
7.5.1. Description
Returns the progress of a job (model creation, model testing or model projection). Possible values are: -1 if the job is still queued, -2 if the job was aborted, a value between 0 and 100 if the job is in progress or 100 if the job was finished. More details about aborted jobs can be discovered with the getLog operation.
7.5.2. SOAP faults
- Missing ticket in request
- Unknown ticket
- Progress unreadable
7.5.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<SOAP-ENV:Envelope xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
SOAP-ENV:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/">
<SOAP-ENV:Body>
<omws:getProgress xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
SOAP-ENV:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ticket xsi:type="xsd:string">FXPAHp</ticket>
</omws:getProgress>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 9: getProgress request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getProgressResponse>
<progress>100</progress>
</omws:getProgressResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 10: getProgress response.
7.6. getLog
7.6.1. Description
Returns the log of a job.
7.6.2. SOAP faults
- Missing ticket in request
- Log unavailable
7.6.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<SOAP-ENV:Envelope xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
SOAP-ENV:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/">
<SOAP-ENV:Body>
<omws:getLog xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
SOAP-ENV:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ticket xsi:type="xsd:string">FXPAHp</ticket>
</omws:getLog>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 11: getLog request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getLogResponse>
<log>
[Info] Creating model
[Info] Reading 2-dimensional occurrence points.
[Info] Using 5 points to find the bioclimatic envelope.
[Info] Finished creating model
[Info] Calculating ROC curve
[Info] Generating 10000 background points
</log>
</omws:getLogResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 12: getLog response.
7.7. getModel
7.7.1. Description
Returns a created model, which includes all parameters that were used to generate the model. Note in example 14 that the <Algorithm> element now includes a <Model> element with the actual result of the modelling process for that particular algorithm. This element contains all necessary information for algorithm predictions given an environmental scenario. Each algorithm has its own unique XML representation.
Model testing and model projection require the <Algorithm> section returned by this operation.
7.7.2. SOAP faults
- Missing ticket in request
- Model unavailable
7.7.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<SOAP-ENV:Envelope xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
SOAP-ENV:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/">
<SOAP-ENV:Body>
<omws:getModel xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
SOAP-ENV:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ticket xsi:type="xsd:string">FXPAHp</ticket>
</omws:getModel>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 13: getModel request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getModelResponse>
<ModelEnvelope xmlns="http://openmodeller.cria.org.br/xml/1.0">
<SerializedModel>
<Sampler>
<Environment NumLayers="2">
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66" IsCategorical="0" Min="-281" Max="310"/>
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/bf441530-04d9-11e1-be50-0800200c9a66" IsCategorical="0" Min="9" Max="211"/>
<Mask Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66"/>
</Environment>
<Presence Label="Test species" Count="5">
<CoordinateSystem>GEOGCS['1924 ellipsoid',DATUM['Not_specified',SPHEROID['International 1924',6378388,297,
AUTHORITY['EPSG','7022']],AUTHORITY['EPSG','6022']],PRIMEM['Greenwich',0,AUTHORITY['EPSG','8901']],UNIT['degree',
0.0174532925199433,AUTHORITY['EPSG','9108']],AUTHORITY['EPSG','4022']]</CoordinateSystem>
<Point Id="1" X="-68.85" Y="-11.15" Sample="250 107"/>
<Point Id="2" X="-67.38" Y="-14.32" Sample="260 114"/>
<Point Id="3" X="-67.15" Y="-15.52" Sample="225 112"/>
<Point Id="4" X="-65.12" Y="-16.73" Sample="254 112"/>
<Point Id="5" X="-63.17" Y="-17.80" Sample="241 103"/>
</Presence>
</Sampler>
<Algorithm Id="BIOCLIM" Version="0.2">
<Parameters>
<Parameter Id="StandardDeviationCutoff" Value="0.8"/>
</Parameters>
<Model>
<Bioclim Mean="246 109.6" StdDev="10.9 3.60" Minimum="225 103" Maximum="260 114"/>
</Model>
</Algorithm>
<Statistics>
<ConfusionMatrix Threshold="0.5" Accuracy="100" OmissionError="0" CommissionError="-100"
TruePositives="5" FalsePositives="0" TrueNegatives="0" FalseNegatives="0"/>
<RocCurve Auc="0.99" NumBackgroundPoints="10000" Points="0 0 0.0087 0.40 0.0087 0.40 0.0087 0.40 0.0087 0.40
0.0087 0.40 0.0087 0.40 0.0087 0.40 0.021 1 0.021 1 0.021 1 0.021 1 0.021 1 0.021 1 0.021 1 1 1"/>
</Statistics>
</SerializedModel>
</ModelEnvelope>
</omws:getModelResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 14: getModel response.
7.8. testModel
7.8.1. Description
Starts a new model test job. This requires an environmental scenario (set of layers including a mask), a set of presence points, an optional set of absence points, a model (<Algorithm> element returned by the createModel operation), and statistics parameters.
Statistics may include confusion matrix and ROC curve. A threshold can be specified for the confusion matrix. In this case, -1 can be used to instruct the server to use the lowest presence threshold (lowest prediction among training points). The "IgnoreAbsences" attribute can be used discard absence points in the calculations, in case they were passed as background points (some algorithms that require background points, like Maxent, have a parameter that allows users to specify the background points through absences).
Two approaches can be used to calculate ROC curves. The first one is the traditional approach which requires presence and absence points. The second was proposed by Phillips et al. (2006) to handle presence-only data using the proportional predicted area. In openModeller this value is approximated by means of background points. When only presences are available, the second approach is always used and the attribute "BackgroundPoints" can be used to specify the number of background points to be generated (default is 10000). Background points can also be specified by the user by means of absences. In this case the attribute "UseAbsencesAsBackground" must be 1. In both approaches the resolution (number of points in the curve) and a maximum accepted ommision can be specified.
This operation is asynchronous. It immediately returns a ticket that must be used to track job progress later.
7.8.2. SOAP faults
- Failed to create ticket
7.8.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<SOAP-ENV:Envelope xmlns:xsi="http://www.w3.org/1999/XMLSchema-instance"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:xsd="http://www.w3.org/1999/XMLSchema"
SOAP-ENV:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/">
<SOAP-ENV:Body>
<omws:testModel xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
SOAP-ENV:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<TestParameters xmlns="http://openmodeller.cria.org.br/xml/1.0">
<Sampler>
<Environment NumLayers="2">
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66" IsCategorical="0"/>
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/bf441530-04d9-11e1-be50-0800200c9a66" IsCategorical="0"/>
<Mask Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66"/>
</Environment>
<Presence Label="Test species">
<CoordinateSystem>GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,
AUTHORITY["EPSG","7030"]],TOWGS84[0,0,0,0,0,0,0],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],
UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9108"]],AXIS["Lat",NORTH],AXIS["Long",EAST],AUTHORITY["EPSG","4326"]]
</CoordinateSystem>
<Point Id="1" X="-67.845739" Y="-11.327340" />
<Point Id="2" X="-69.549969" Y="-12.350801" />
</Presence>
<Absence Label="Test species">
<CoordinateSystem>GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,
AUTHORITY["EPSG","7030"]],TOWGS84[0,0,0,0,0,0,0],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],
UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9108"]],AXIS["Lat",NORTH],AXIS["Long",EAST],AUTHORITY["EPSG","4326"]]
</CoordinateSystem>
<Point Id="3" X="-68.245959" Y="-12.060403" />
<Point Id="4" X="-74.534959" Y="-15.340201" />
</Absence>
</Sampler>
<Algorithm Id="BIOCLIM" Version="0.2">
<Parameters>
<Parameter Id="StandardDeviationCutoff" Value="0.8"/>
</Parameters>
<Model>
<Bioclim Mean="246 109.6" StdDev="10.9 3.60" Minimum="225 103" Maximum="260 114"/>
</Model>
</Algorithm>
<Statistics>
<ConfusionMatrix Threshold="0.5" IgnoreAbsences="0"/>
<RocCurve Resolution="15" MaxOmission="1.0" UseAbsencesAsBackground="0"/>
</Statistics>
</TestParameters>
</omws:testModel>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 15: testModel request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:testModelResponse>
<ticket>9HhPVc</ticket>
</omws:testModelResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 16: testModel response.
7.9. getTestResult
7.9.1. Description
Returns the result of a model test. Points of the ROC curve are serialized as a sequence of pairs (x y) separated by a space.
7.9.2. SOAP faults
- Missing ticket in request
- Test result unavailable
7.9.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:getTestResult xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
soap:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ticket xsi:type="xsd:string">9HhPVc</ticket>
</omws:getTestResult>
</soap:Body>
</soap:Envelope>
Example 17: getTestResult request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:testResponse>
<TestResultEnvelope xmlns="http://openmodeller.cria.org.br/xml/1.0">
<Statistics>
<ConfusionMatrix Threshold="0.5" Accuracy="75" OmissionError="0" CommissionError="50"
TruePositives="2" FalsePositives="0" TrueNegatives="1" FalseNegatives="1"/>
<RocCurve Auc="0.875" Points="0 0 0 0.5 0 0.5 0 0.5 0 0.5 0 0.5 0 0.5 0 0.5 0.5 1 0.5 1 0.5 1 0.5 1 0.5 1
0.5 1 0.5 1 1 1 1 1"/>
</Statistics>
</TestResultEnvelope>
</omws:testResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 18: getTestResult response.
7.10. projectModel
7.10.1. Description
Starts a new model projection job. This includes a model (<Algorithm> section returned by the createModel operation), an environmental scenario (set of layers with an output mask indicating the extent of the projection) and additional output parameters. If the environmental scenario is the same as the one used in model creation, this is known as a native projection. If the scenario is different, the number of layers must be the same and the corresponding variables must also be in the same order used for model creation.
Output parameters include a template layer whose spatial resolution and spatial reference system will be used to create the distribution map, and a file type with the possible values:
| GreyTiff | Grey scale GeoTiff [0,255] |
| GreyTiff100 | Grey scale GeoTiff [0,100] |
| FloatingTiff | Floating point GeoTiff |
| GreyBMP | Grey scale BMP |
| FloatingHFA | Floating point Erdas Imagine |
| ByteHFA | Byte Erdas Imagine [0,100] |
| ByteASC | Byte ARC/Info ASCII grid [0,100] |
| FloatingASC | Floating point ARC/Info ASCII grid |
An optional <Statistics> section can be used to specify the threshold to calculate the number of cells considered suitable for the species occurrence. This information can be retrieved through the getProjectionMetadata operation.
This operation is asynchronous. It immediately returns a ticket that must be used to track job progress later.
7.10.2. SOAP faults
- Failed to create ticket
7.10.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:projectModel xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
soap:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ProjectionParameters xmlns="http://openmodeller.cria.org.br/xml/1.0">
<Algorithm Id="BIOCLIM" Version="0.2">
<Parameters>
<Parameter Id="StandardDeviationCutoff" Value="0.8"/>
</Parameters>
<Model>
<Bioclim Mean="246 109.6" StdDev="10.9 3.60" Minimum="225 103" Maximum="260 114"/>
</Model>
</Algorithm>
<Environment NumLayers="2">
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66"/>
<Map Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/bf441530-04d9-11e1-be50-0800200c9a66"/>
<Mask Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66"/>
</Environment>
<OutputParameters FileType="ByteHFA">
<TemplateLayer Id="/layers/en/terrestrial/climate/global/worldclim/present/bioclim/10arc-minutes/14aecd40-04d9-11e1-be50-0800200c9a66"/>
</OutputParameters>
<Statistics>
<AreaStatistics PredictionThreshold="0.5"/>
</Statistics>
</ProjectionParameters>
</omws:projectModel>
</soap:Body>
</soap:Envelope>
Example 19: projectModel request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:projectModelResponse>
<ticket>Umz5UR</ticket>
</omws:projectModelResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 20: projectModel response.
7.11. getLayerAsUrl
7.11.1. Description
Returns the URL of a projected model.
7.11.2. SOAP faults
- Missing id in request
- Layer unavailable
7.11.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:getLayerAsUrl xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
soap:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<id xsi:type="xsd:string">xvQD6d</id>
</omws:getLayerAsUrl>
</soap:Body>
</soap:Envelope>
Example 21: getLayerAsUrl request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getLayerAsUrlResponse>
<url>http://modeller.cria.org.br/maps/xvQD6d.img</url>
</omws:getLayerAsUrlResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 22: getLayerAsUrl response.
7.12. getProjectionMetadata
7.12.1. Description
Returns metadata about a model projection given a ticket, including the total number of cells on the map, the number of cells predicted as suitable for the species and the threshold used to calculate these values.
7.12.2. SOAP faults
- Missing ticket in request
- Projection unavailable
- Projection unreadable
- Metadata unavailable
7.12.3. Examples
<?xml version="1.0" encoding="iso-8859-1"?>
<soap:Envelope xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:soapenc="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
soap:encodingStyle="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:soap="http://schemas.xmlsoap.org/soap/envelope/">
<soap:Body>
<omws:getProjectionMetadata xmlns:omws="http://openmodeller.cria.org.br/ws/1.0"
soap:encodingStyle="http://xml.apache.org/xml-soap/literalxml">
<ticket xsi:type="xsd:string">xvQD6d</ticket>
</omws:getProjectionMetadata>
</soap:Body>
</soap:Envelope>
Example 23: getProjectionMetadata request.
<?xml version="1.0" encoding="UTF-8"?>
<SOAP-ENV:Envelope xmlns:SOAP-ENV="http://schemas.xmlsoap.org/soap/envelope/"
xmlns:SOAP-ENC="http://schemas.xmlsoap.org/soap/encoding/"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:xsd="http://www.w3.org/2001/XMLSchema"
xmlns:om="http://openmodeller.cria.org.br/xml/1.0"
xmlns:omws="http://openmodeller.cria.org.br/ws/1.0">
<SOAP-ENV:Body>
<omws:getProjectionMetadataResponse FileSize="81786">
<ProjectionEnvelope xmlns="http://openmodeller.cria.org.br/xml/1.0">
<AreaStatistics TotalCells="585176" CellsPredicted="13064" PredictionThreshold="0.5"/>
</ProjectionEnvelope>
</omws:getProjectionMetadataResponse>
</SOAP-ENV:Body>
</SOAP-ENV:Envelope>
Example 24: getProjectionMetadata response.
8. References
Phillips, S.J., Anderson, R.P., and Schapire, R.E., 2006. Maximum entropy modeling of species geographic distributions. Ecological Modelling 190: 231–259.
Muñoz, M.E.S., Giovanni, R., Siqueira, M.F., Sutton, T., Brewer, P., Pereira, R.S., Canhos, D.A.L., Canhos, V.P., 2011. openModeller: a generic approach to species’ potential distribution modelling. Geoinformatica, 15: 111-135.