Overview
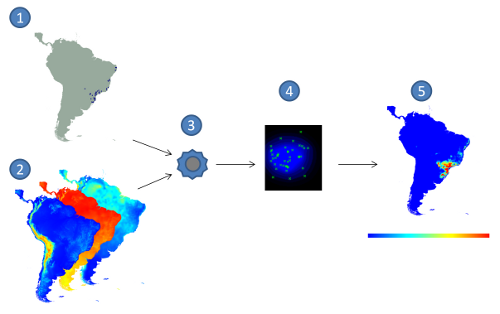
Figure 1: Correlative approach for Ecological Niche Modelling. (1) species occurrence points,
(2) environmental layers, (3) modelling algorithm, (4) model in the environmental space,
(5) model projection, with red indicating higher suitability values and blue the lower ones.
Ecological Niche Modelling
Ecological Niche Modelling (ENM) is a widely used approach to predict and to understand the distribution of species. An ecological niche can be seen as the set of ecological requirements for a certain species to survive and maintain viable populations over the time (Grinnell, 1917). In most cases, ENMs are generated by relating locations where the species is known to occur with environmental variables that may influence its distribution, and then applying an algorithm to create a model (figure 1). This method is known as correlative approach (Soberón & Peterson, 2005). The resulting model basically tries to find a representation of the conditions that are suitable for the species. Such models can be projected into different geographical regions under different environmental scenarios, making it possible to predict the impact of climate changes on biodiversity, prevent the spread of invasive species, identify geographical and ecological aspects of disease transmission, help in conservation planning, guide field surveys, among many other uses (Peterson et. al. 2011).
openModeller
openModeller (Muñoz et. al. 2009) is an open source framework written in C++ to perform ecological niche modelling tasks. When it was created at CRIA more than 10 years ago, it was seen as a natural step considering CRIA's previous experience in developing and using Desktop Garp in a joint effort with the University of Kansas. Desktop Garp is a typical niche modelling software: it uses a single algorithm (GARP), supports a single input format for environmental layers, requires all layers to have the same extent and resolution, was developed having a single interface in mind, runs only in one platform and the source code is not readily available. The same applies to many other software options in this area. Therefore, the main motivation for creating openModeller was to support a larger number of input formats, save users from having to learn different software in order to use different algorithms, save users from the tedious and sometimes unnecessary tasks of clipping and cropping environmental layers, allow multiple interfaces to be created on top of the same framework, run in the most popular platforms and be completely open source.

Figure 2: openModeller architecture.
A modular architecture (figure 2) developed as a multi-platform framework was used to overcome all these issues. Different formats for species occurrence data and environmental data are handled by specific drivers, different spatial resolutions and spatial reference systems can be automatically harmonised by the framework, which also includes several algorithms implemented as plugins. The framework was developed without having any particular interface in mind, therefore facilitating the task of creating new interfaces. Currently there are command-line, Desktop and Web Service interfaces available, including a module for the Python programming language. The following algorithms are included in openModeller: GARP Best Subsets (two different implementations), Maxent, ENFA, Suppor Vector Machines (three different types), Random Forests, Artificial Neural Networks, AquaMaps, CSM, Environmental Distance algorithms, Bioclim, among others.
The framework makes use of other C/C++ libraries, such as GDAL (Geospatial Data Abstraction Library), PROJ.4 (Cartographic Projections Library), Expat (XML Parser), libcURL and GSL (GNU Scientific Library). Optional additional libraries can be used depending on the desired functionality. For developers, compilation is based on CMake and the code is known to compile and run on GNU/Linux, Windows, Mac OSX and *BSD platforms using 32bit or 64bit x86 processors.
The framework contains two drivers that can both read (environmental layers) and write (projected models) raster data. One of them is based on GDAL, which allows all GDAL supported formats to be used, including remote rasters available through WCS providers. The other driver was created to read and write rasters stored on TerraLib databases.
Another kind of driver is used to read species occurrence points, with separate implementations for each supported data source: local TAB-delimited files, GBIF REST service, speciesLink web service and TerraLib databases. The library Proj.4 is used to convert between different georeferencing systems and projections.
More details about openModeller can be found in Muñoz et. al. (2009).
References
Grinnell, J. (1917) Field tests of theories concerning distributional control. American Naturalist, 51: 115-128.
Muñoz, M.E.S., Giovanni, R., Siqueira, M.F., Sutton, T., Brewer, P., Pereira, R.S., Canhos, D.A.L. & Canhos, V.P. (2009) "openModeller: a generic approach to species' potential distribution modelling". GeoInformatica. DOI: 10.1007/s10707-009-0090-7
Peterson, A.T., Soberón, J., Pearson, R.G., Anderson, R.P., Martinez-Meyer, E., Nakamura, M. & Araújo, M.B. (2011) Ecological niches and geographic distributions. Princeton University Press. ISBN 978-0-691-13688-2.
Soberón, J. & Peterson, A.T. (2005) Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodiversity Informatics, 2: 1-10.
Last updated on 10 March 2014

